Challenges and pitfalls in the use of partitioned gene counts for homoeologous gene expression and co-expression network analyses
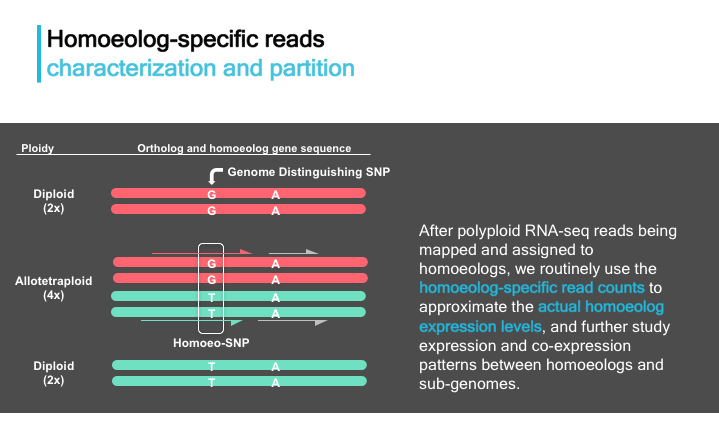
As shown below in an allotetraploid genome, homoeologs refers to the duplicated gene pairs originated from its parental diploids. Only sequencing reads containing homoeolog distinguishing SNPs can be assigned to either homoeolog genes, while some reads like the two grey ones here remain indistinguishable.
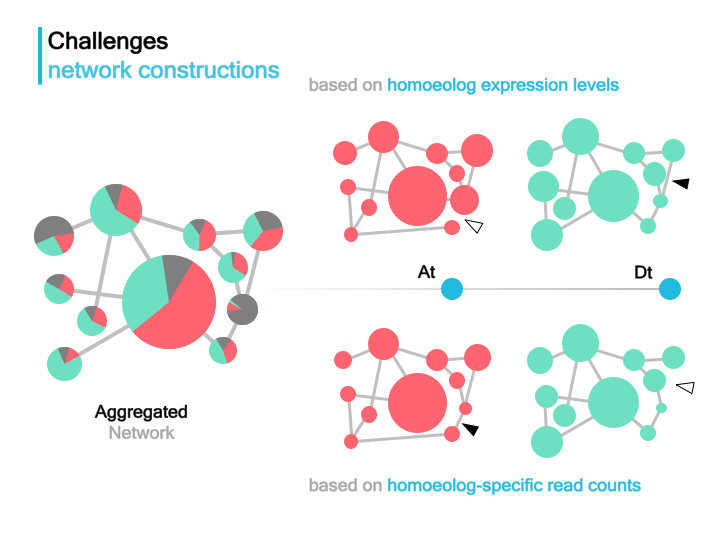
The resulted networks will show differences from the expected network, as indicated by arrows.
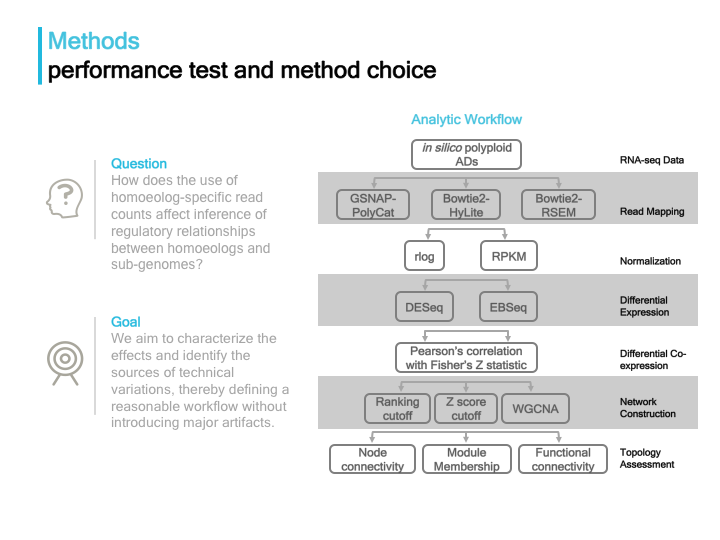
Applying the analysitic workflow below, we tested different methods at multiple stages as a possible source of technical variance, and also compared their performances.
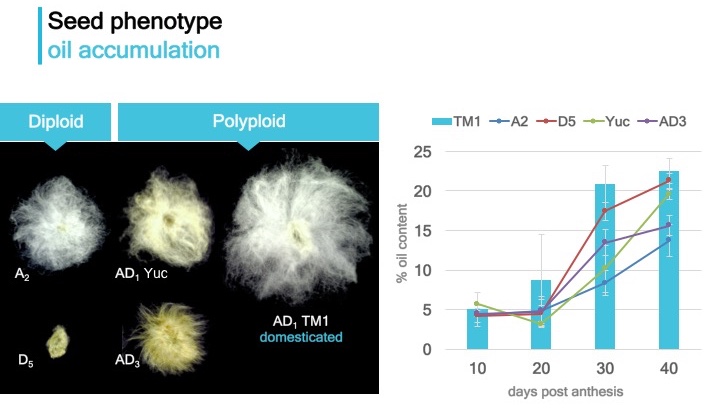
Co-expression gene network of cottonseed development
To gain insight into phenotypic diversification in cotton seeds, we conducted co-expression network analysis of developing seeds from diploid and allopolyploid cotton species and explored network properties.
Two different conceptual approaches were explored to help understand network conservation and divergence at various evolutionary timescales and across ploidy levels..
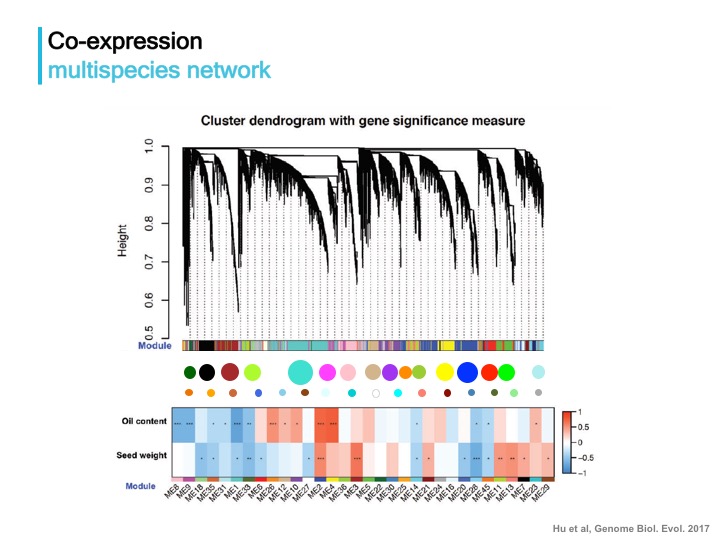
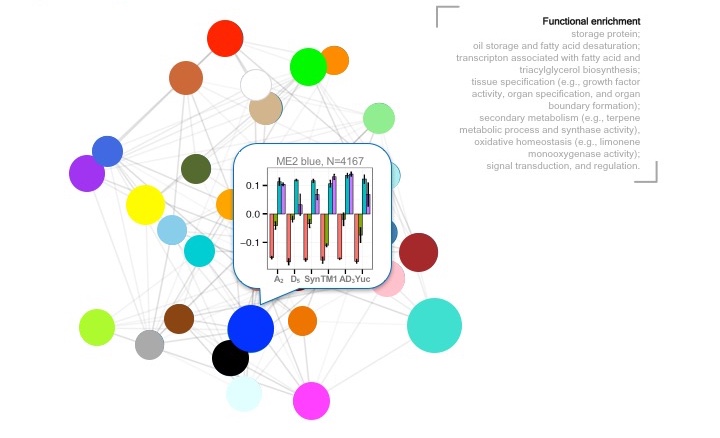
Approach One: construct a multispecies coexpression network to provide a global view of coexpression network topology among species, followed by identifying key modules associated with devlopmental dynamics and phenotypic diversification
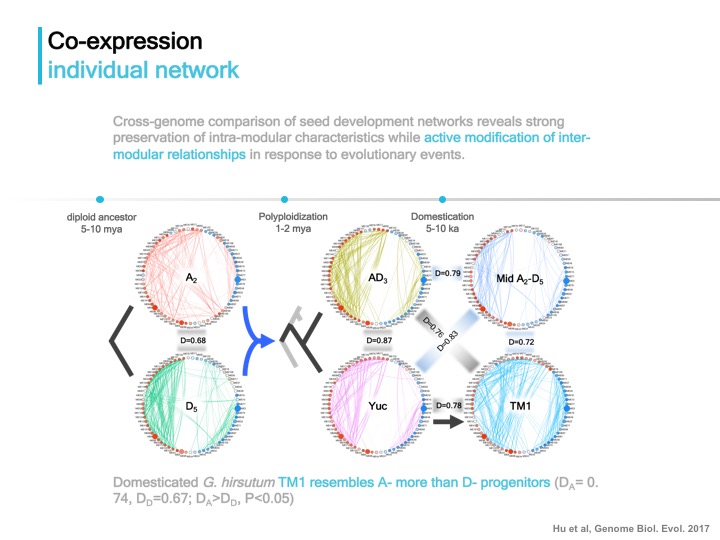
Approach Two: construct individual coexpression networks for comparison to reveal topological changes in response to evolutionary events
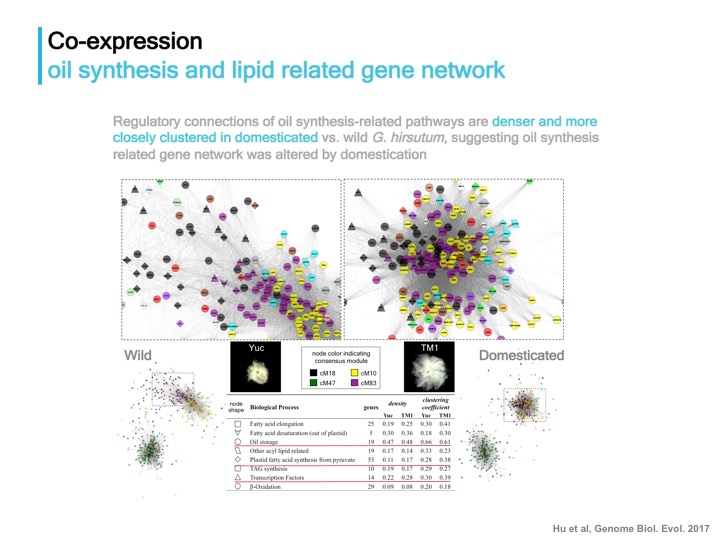
Homoeolog gene co-expression network implicates biased recruitment of regulatory architecture of seed development
Instead of using the aggregated expression of homoeolog gene pairs, we now constructed allopolyploid co-expression networks based on homoeolog-specific expressions. This approach allows direct comparisons of sub-genome networks against their corresponding diploid parental networks. In addition to revealing topological modificantions of the “redundant” duplicated seed network following allopolyploidization, this work aims to explore the assymetric evolution of parental regulatory connections underlying phenotypic evolution and domestication.
Stay tuned!