This project aims to enhance our understanding of the genotype-to-phenotype connection in polyploid genomes, by characterizing both chromatin architecture and gene activities, and their responses to environmental perturbations such as salt and drought stress. We hypothesize that the dynamics of nucleosome organization and chromatin accessibility drive gene expression responses to speciation and adaptation. Using the cotton genus (Gossypium) as our model and the powerful lens of evolutionary biology, we will globally determine nucleosome positioning, the regulatory control of duplicated genes, and genome-wide gene regulatory networks. The goals are to:
- Characterize genome-wide nucleosome organization and chromatin accessibility in closely related diploids and their allopolyploid descendants, thereby providing insight into the evolutionary divergence of cis-regulatory landscapes and how these are altered by genome merger and doubling.
- Determine the relationships between chromatin architecture and gene expression evolution accompanying allopolyploidization, thereby providing insight into the poorly understood phenomenon of duplicate gene coregulation, and the origin of regulatory novelty
- Elucidate the cis-regulatory landscapes in stress-responsive regulatory gene networks in diploids and polyploids, thereby facilitating insight into polyploidy and stress biology.
Nucleosome organization and chromatin accessibility
Genomes of eukaryotes are organized into chromatin, comprising a complex of genetic material and various proteins. The fundamental structural units of chromatin are nucleosomes. The positioning and stability of nucleosomes regulate the exposure of naked DNA for gene transcription and other DNA-dependent processes including replication, repair and recombination. Nucleosome positioning may be conceptualized as an emergent outcome of the myriad epigenetic modifications of histones and DNA that collectively control gene expression. Over the past decade, the implementation of high-throughput techniques have enabled genome-wide mapping of nucleosome occupancy in model organisms, including several plant species (e.g., arabidopsis, rice and maize). Despite the strong association observed between chromatin structure and gene expression, little is understood about how these relationships are altered during evolution.
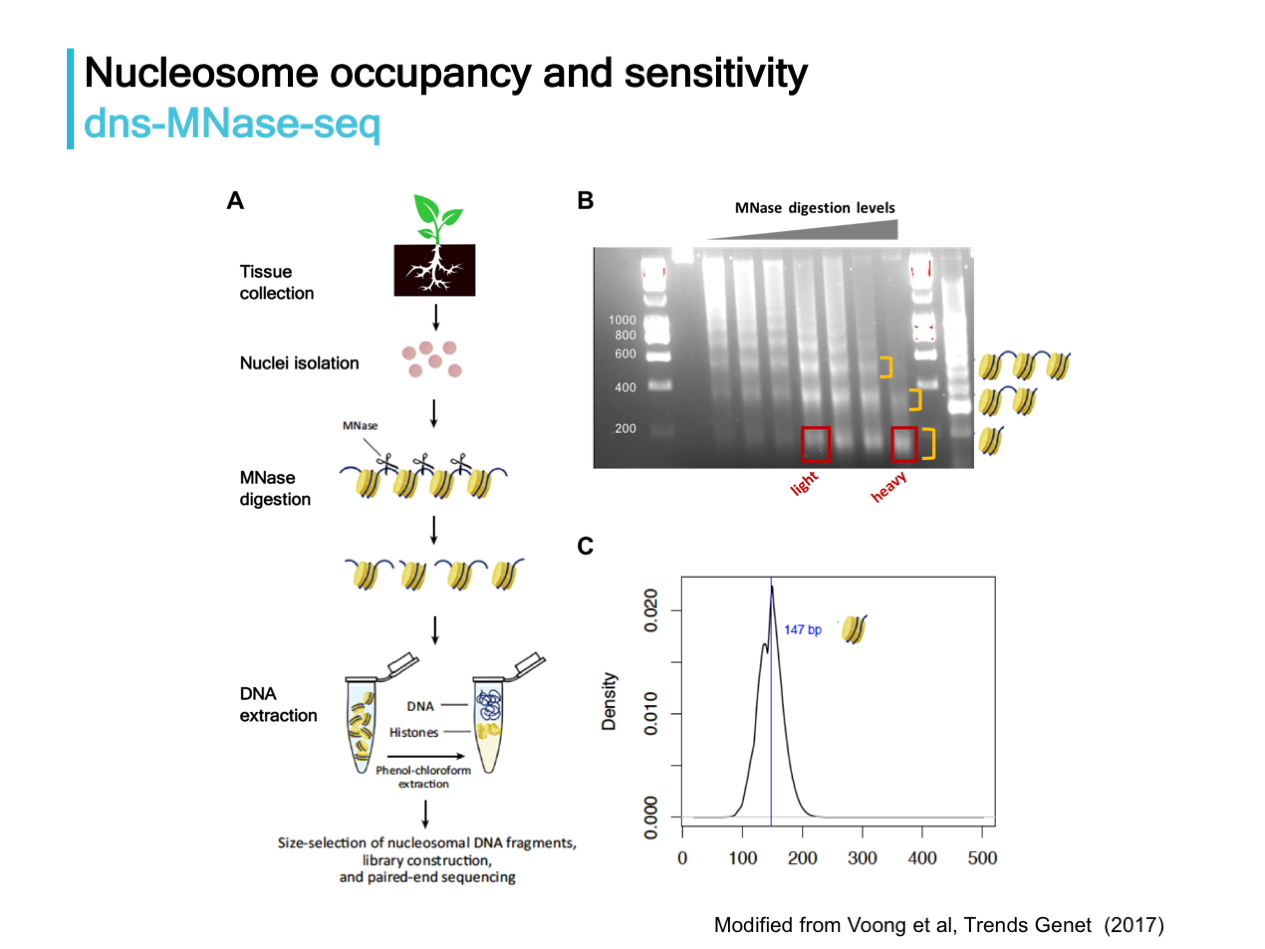
Differential nuclease sensitivity profiling (dns-MNase-seq) is a powerful approach for chromatin profiling, as shown for maize (Vera et al. 2014), not only for genome-wide profiling of nucleosome positioning and stability, but also for characterizing functional cis-regulatory elements. Here we plan to apply dns-MNase-seq to cotton tissues at different ploidy levels to characterize nucleosome dynamics in conjunction with expression data to elucidate and quantify gene regulatory divergence over evolutionary time and in response to environmental cues (e.g., salt and drought).