This research aims to develop our understanding how evolutionary history, including natural polyploidization and human-mediated domestication, sculpted cotton proteomes. These in-depth functional genomic data are also expected to provide us new opportunities for cotton improvement. Our specific research objectives are:
- To develop technology and tools for describing and studying the cotton fiber and seed proteomes.
- To describe the cotton proteome from the standpoint of fiber development, which will allow us to assess the changes that accompany fiber evolution and domestication, and how this correlates with existing information on the transcriptome.
- To understand how the proteome responds to genome doubling; that is, what is novel about polyploid cotton fiber and seed relative to that of its antecedent diploids?
- To detail proteomic consequences of cotton fiber evolution and domestication; for example, to catalog the key proteins associated with and therefore possibly responsible for phenotype changes and important traits relevant to crop improvement.
Using proteomic approaches including 2-DE gel electrophoresis, iTRAQ and label-free , comparative experiments were conducted on developing fibers and seeds in diploid and allopolyploid cotton species.
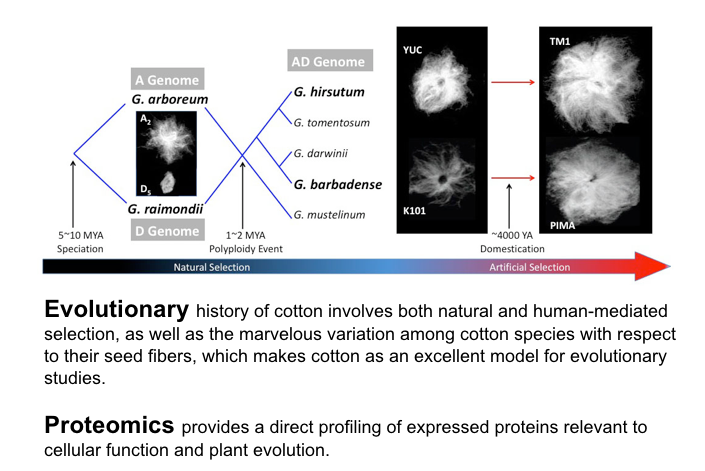
Cotton proteomes of mature seeds 1 and developing fibers 2 were examined to understand the genome-wide proteomic changes accompanying allopolyploidization. Interspecific comparisons of protein composition and expression level among diploid and polyploid cotton species revealed that the allopolyploid cotton display asymmetric proteomic divergence with respect to the diploid progenitors, which is detailed by describing important phenomena of non-additive expression, expression level dominance and homoeolog expression bias.
To understand the genome-wide expression changes associated with domestication, the fiber proteomes represented by four representative developmental stages were studied for two important crop species - G. hirsutum 3 and G. barbadense 4, using paired wild and domesticated accessions. By contrasting the fiber proteomes and developmental dynamics between wild and domesticated accessions for each species, we were able to characterize the key features of global protein pattern change corresponding to the domestication processes. Furthermore, we identified a number of proteins differentially expressed during fiber development and altered by domestication, as candidate proteins and metabolic processes for functional analyses that may yield insight into domestication and future cotton improvement.
Reference
1. Hu, G., N. L. Houston, D. Pathak, L. Schmidt, J. Thelen and J. F. Wendel. 2011. Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics 189: 1103-1115. PDF
2. Hu, G., J. Koh, M. J. Yoo, S. Chen, and J. F. Wendel. 2015. Gene-expression novelty in allopolyploid cotton: a proteomic perspective. Genetics 200: 91-104. PDF
3. Hu, G., J. Koh, M. J. Yoo, D. Pathak, S. Chen, and J. F. Wendel. 2014. Proteomics profiling of fiber development and domestication in upland cotton (Gossypium hirsutum L.). Planta 240: 1237-1251. PDF
4. Hu, G., J. Koh, M. Yoo, K. Grupp, S. Chen, and J. F. Wendel. 2013. Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense. New Phytologist 200: 570–582 PDF